Translate this page into:
A case of Philadelphia chromosome-positive acute myeloid leukaemia with missense mutation R132c (c.394 c>t) and single nucleotide polymorphism rs11554137(G105G) in isocitrate dehydrogenase 1 gene
Corresponding Author:
Akanksha Agarwal
Department of Pathology, King George Medical University, Lucknow, Uttar Pradesh
India
agarwal.akanksha8@gmail.com
| How to cite this article: Agarwal A, Jain M, Tewari S, Goel M, Kumar A. A case of Philadelphia chromosome-positive acute myeloid leukaemia with missense mutation R132c (c.394 c>t) and single nucleotide polymorphism rs11554137(G105G) in isocitrate dehydrogenase 1 gene. Natl Med J India 2020;33:146-148 |
Abstract
Acute myeloid leukaemia (AML) is a heterogeneous disease due to its variable clinical, morphological and genetic features. New chromosomal and molecular abnormalities are being studied to evaluate novel treatment options and prognostic implications. We report a patient with AML who was Philadelphia chromosome-positive along with IDH1R132 mutation and SNP rs11554137(IDH1105GGT). Due to limited reports regarding these aberrations in patients with AML, there is no consensus regarding their prognostic implications. To the best of our knowledge, the presence of Philadelphia chromosome, IDH1R132 and SNP rs11554137 in a single AML patient with good prognosis is a novel findingIntroduction
Acute myeloid leukaemia (AML) is associated with high morbidity and mortality, has a poor prognosis with 5-year survival of about 25%.[1],[2] Studies have revealed that pathogenesis of AML involves the abnormal proliferation and differentiation of clonal population of myeloid stem cells. WHO has emphasized the importance of bone marrow morphology, cytochemistry, flowcytometry, cytogenetics and molecular analyses for the diagnosis of AML.
Approximately 50%–60% of de novo patients of AML are associated with cytogenetic abnormalities.[3] WHO has categorized these chromosomal aberrations into various cytogenetic risk groups. Philadelphia (Ph) chromosome (t[9;22])—a hallmark of chronic myeloid leukaemia (CML) has been seen in a few patients of AML. This translocation is known to produce a fusion protein affecting the tyrosine kinase pathway leading to increased proliferation, decreased differentiation and apoptosis. Although t(9;22) has been included as a provisional entity in WHO 2016 AML classification, its diagnostic criteria and treatment protocols are unclear due to the limited number of patients reported.
Studies have elucidated the importance of gene mutations, which are identified in more than 97% of patients with AML.[4],[5] WHO has classified these mutations into three classes depending on their prognostic significance. Isocitrate dehydrogenase 1 (IDH1) R132 mutation has been associated with 7%–10% of patients with AML with a variable prognosis.[6] It leads to impaired cell differentiation along various lineages and to tumour development along with other cancer genes and tumour-causing factors. A silent SNP at codon 105 of exon 4 has been reported in patients of AML and is proposed to be associated with poor prognosis.[6] We report a patient with concurrent t(9;22), IDH1R132 mutation and SNP rs11554137 (IDH1105 GGT) with good remission and survival.
The Case
A 33-year-old female presented in 2017, with history of mild to moderate fever for 3 months. Systemic examination was unremarkable (no organomegaly or lymphadenopathy). Her complete blood count showed a haemoglobin of 7.9 g/dl, white blood cell count 26.5×109/L and platelet count of 350×109/L. Peripheral smear examination revealed majority of cells (82%) were medium- to large-sized blast cells with moderately fine chromatin, 1–2 nucleoli and moderate amount of cytoplasm. Cytochemical stain for myeloperoxidase was positive and for periodic acid–Schiff was negative. Bone marrow showed increased cellularity with 73% blast population with the same morphology as described in the peripheral smear. There was no Auer rod or dysplasia. Bone marrow examination was consistent with the diagnosis of AML. Flow cytometric immuno-phenotyping of blood sample showed myeloid blasts (CD34, HLA DR, CD13, CD33, cMPO were positive and CD2, CD7, CD10 and CD 38 were negative). Cytogenetic analysis showed a balanced translocation between long arm of chromosome 9 and 22 between the regions q34 and q11.2, respectively. Reverse transcriptase-polymerase chain reaction (PCR) studies showed BCR-ABL rearrangement showing major translocation (p210). PCR followed by sequencing showed IDH1R132 mutation and SNP G105. NPM1, FLT3 TKD/ ITD mutation was not detected [Figure - 1] to [Figure - 3]. The patient was treated with (7+3) regime comprising high-dose cytosine arabinose and daunorubicin along with imatinib. Repeat bone marrow examination after 1 month showed complete remission with bone marrow blasts <1%. Repeat quantitative analysis of BCR-ABL1 after 12 months was 1% (BCR-ABL1 [IS]). Since then, the patient is on remission consolidation therapy comprising high-dose cytarabine along with imatinib and regular follow-up till date. There was no recurrence of disease till 2018.
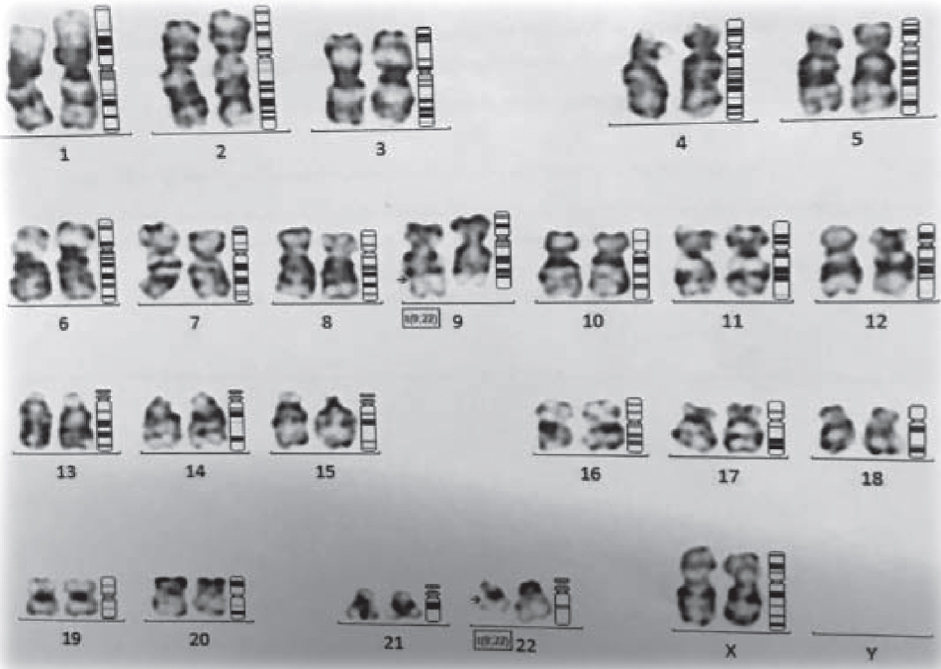 |
| Figure 1: G-banded karyotype of bone marrow cells showing 46, X, t(9;22) (q34;q11.2) |
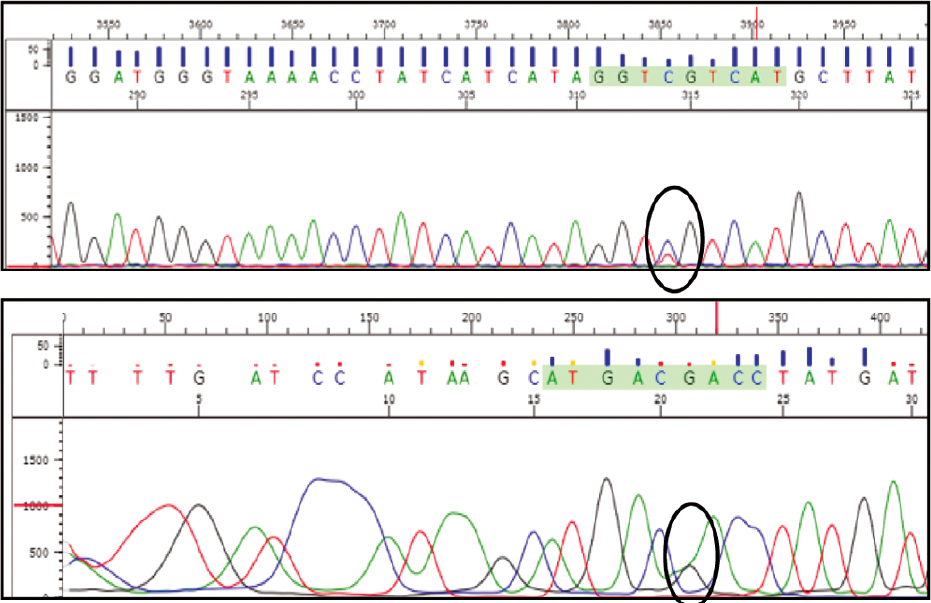 |
| Figure 2: Electropherograms of isocitrate dehydrogenase 1 exon 4 with heterozygous missense mutation R132C |
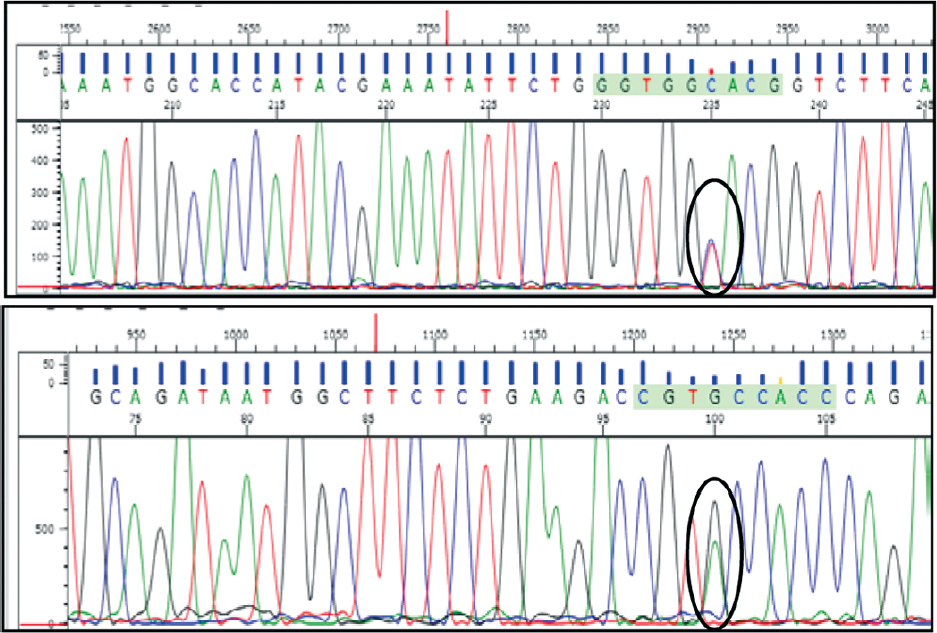 |
| Figure 3: Electropherograms of isocitrate dehydrogenase 1 exon 4 with heterozygous polymorphism G105G (c.315 C>T) |
Discussion
We report a patient with coexisting t(9;22), IDH1R132 mutation and SNP IDHG105. Lack of data on these entities make this case interesting. Furthermore, all these events, whenever reported individually, have been associated with poor prognosis; however, our patient has been under treatment for over 1 year and is in complete cytological remission.
AML has uncontrolled proliferation and minimal differentiation due to acquired genetic alterations. Known cytogenetic aberrations have been used for the classification of AML into various cytogenetic risk groups. 7+3 regime involving cytosine arabinose and daunorubicin has been the cornerstone of treatment of patients with AML. Prognosis of AML is variable with some patients having long-term disease-free survival while others having more aggressive disease. The 5-year survival is 35%–40% but in the older population it is <15%.[7]
Philadelphia chromosome is observed in CML, ALL and mixed phenotypic acute lenkaemia. Incidence of Ph+ in de novo AML is 0.5%–3%[8] and is associated with a poor prognosis. This has been included as a provisional entity in the WHO 2016 classification. Although difficult to differentiate CML-BC and Ph+ AML, lack of splenomegaly and low levels of basophils support the diagnosis of Ph+ AML. Our patient had no additional chromosomal abnormality such as trisomy 8,19 or inversion 17q, which have been commonly observed in Ph+ CML. ABL1 mutation, reported frequently in CML-BC, was also not detected. BCR-ABL1 fusion protein affects the tyrosine kinase pathway causing increased proliferation, decreased differentiation and apoptosis. Involvement of tyrosine kinase brings in the importance of molecular targeted therapy, i.e. tyrosine kinase inhibitors (TKI). These are known to provide good treatment response and prognosis in tumours involving this pathway. TKI in combination with 7+3 regime should be a better treatment plan for patients with Ph+ AML. Our patient was given imatinib along with standard AML treatment and is in complete cytological remission and event-free follow-up.
Mutation in IDH1 was first analysed by exome resequencing in patients with colorectal cancer. Subsequently, IDH1 and IDH2 mutations were studied in various other malignancies. In the 2016 WHO classification of central nervous system tumours, IDH mutated and wild-type entities have been included in the diagnostic criteria of gliomas.[9] Normal IDH enzymes are involved in many cellular processes such as mitochondrial oxidative phosphorylation, glutamine metabolism, lipogenesis, glucose sensing and regulation of cellular redox status. NADPH generated by wild-type IDH1 and IDH2 during cellular metabolism is a cellular-reducing agent required for detoxification processes. The normal metabolite α ketoglutarate (aKG) acts as a co-factor in a wide range of cellular processes such as hypoxia, angiogenesis, maturation of collagens of extracellular matrix and regulation of epigenetics.[10]
Mutations of the arginine residue in the active site (R132 in IDH1 and R140, R172 in IDH2) prevent normal function of the IDH enzyme and result in the formation of a rare oncometabolite 2D hydroxyl glutarate (D2HG).[11] D2HG competitively inhibits aKG dependent dioxygenases (TET DNA hydroxylases, Jumonji C domain containing histone demethylase and DNA methyl-transferase) leading to hypermethylation of CpG islands in the promoter region. Hypermethylated states due to these mutations lead to blocked haematopetic stem cell/progenitor differentiation, myeloid skewing and eventually to myeloid neoplasms.[12] IDH mutation is present in 5%–15% of patients with cytogenetically normal (CN)-AML. In single nucleotide polymorphism, the single nucleotide change does not affect the amino acid sequence; as a result, the function of protein encoding remains the same. SNP rs11554137 (IDH1105GGT) is prevalent in around 10% of AML cases.[6] This SNP alters RNA stability, folding, splicing, tRNA selection or binding of non-coding RNAs leading to increased IDH1 mRNA. As IDH1 protein level increases, it affects intra-cellular NADPH metabolism that may lead to tumourigenesis.[13] Although the role of IDH1G105 in tumour development is not understood, it has been associated with poor prognosis in patients with CN-AML and glioma. The pathophysiology of IDH can be associated with hypermethylation. Consequently, the discovery of IDH1/2 mutations has resulted in various therapeutic approaches (hypomethylating agents, IDH mutant enzyme inhibitors, immunotherapy and BCL2 inhibition) aimed at either restoring the normal IDH1 and 2 or at blocking D2HG production. Few drugs such as AG-120 and AG-221 are undergoing trials.[14]
Conclusion
This patient is unique in that there are few reported instances of AML with Ph+ (t[9;22]). IDH1R132 mutation has been reported in CN-AML cases and is thought to have a poor prognosis;[16] but, our patient has cytogenetic aberration, IDH1R132 mutation and is showing good treatment response. To the best of our knowledge, there are few reported cases of IDHR132 mutation in AML with cytogenetic aberrations. There are reported cases of AML with SNP IDH1G105, and these also have a poor prognosis. Our patient has coexistent IDH1R132 mutation and SNP IDH1G105 along with Ph+ (t[9;22]) with event-free disease remission phase. This highlights the importance of targeted molecular therapy or designer drugs against a specific protein or molecule. A course of imatinib along with standard AML treatment protocol has possibly led to better prognosis in this patient.
Conflicts of interest. None declared
| 1. | Swerdlow S, Campo E, Harris N, Jaffe E, Pileri S. WHO classifications of tumours of hematopoietic and lymphoid tissues. 4th ed. Lyon:IARC; 2008. [Google Scholar] |
| 2. | Leukemia-Acute Myeloid-AML: Statistics. Cancer. Net.; 2016. [Google Scholar] |
| 3. | Mrózek K, Radmacher MD, Bloomfield CD, Marcucci G. Molecular signatures in acute myeloid leukemia. Curr Opin Hematol 2009;16:64–9. [Google Scholar] |
| 4. | De Kouchkovsky I, Abdul-Hay M. Acute myeloid leukemia: A comprehensive review and 2016 update. Blood Cancer J 2016;6:e441. [Google Scholar] |
| 5. | Molenaar RJ, Thota S, Nagata Y, Patel B, Clemente M, Przychodzen B, et al. Clinical and biological implications of ancestral and non-ancestral IDH1 and IDH2 mutations in myeloid neoplasms. Leukemia 2015;29:2134–42. [Google Scholar] |
| 6. | Wagner K, Damm F, Göhring G, Görlich K, Heuser M, Schäfer I, et al. Impact of IDH1 R132 mutations and an IDH1 single nucleotide polymorphism in cytogenetically normal acute myeloid leukemia: SNP rs11554137 is an adverse prognostic factor. J Clin Oncol 2010;28:2356–64. [Google Scholar] |
| 7. | Stone RM. The difficult problem of acute myeloid leukemia in the older adult. CA Cancer J Clin 2002;52:363–71. [Google Scholar] |
| 8. | Konoplev S, Yin CC, Kornblau SM, Kantarjian HM, Konopleva M, Andreeff M, et al. Molecular characterization of de novo Philadelphia chromosome-positive acute myeloid leukemia. Leuk Lymphoma 2013;54:138–44. [Google Scholar] |
| 9. | Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol 2016;131:803–20. [Google Scholar] |
| 10. | Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer 2012;12:599–612. [Google Scholar] |
| 11. | Yang H, Ye D, Guan KL, Xiong Y. IDH1 and IDH2 mutations in tumorigenesis: Mechanistic insights and clinical perspectives. Clin Cancer Res 2012;18:5562–71. [Google Scholar] |
| 12. | Cazzola M. IDH1 and IDH2 mutations in myeloid neoplasms––Novel paradigms and clinical implications. Haematologica 2010;95:1623–7. [Google Scholar] |
| 13. | Sanson M, Marie Y, Paris S, Idbaih A, Laffaire J, Ducray F, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol 2009;27:4150–4. [Google Scholar] |
| 14. | Mondesir J, Willekens C, Touat M, de Botton S. IDH1 and IDH2 mutations as novel therapeutic targets: Current perspectives. J Blood Med 2016;7:171–80. [Google Scholar] |
| 15. | Singh MK, Gupta R, Rahman K, Yadav G, Sharma A, Nityanand S, et al. De novo Philadelphia positive acute myeloid leukemia with extensive basophilia: A diagnostic dilemma. Indian J Hematol Blood Transfus 2016;32:86–8. [Google Scholar] |
| 16. | Feng JH, Guo XP, Chen YY, Wang ZJ, Cheng YP, Tang YM, et al. Prognostic significance of IDH1 mutations in acute myeloid leukemia: A meta-analysis. Am J Blood Res 2012;2:254–64. [Google Scholar] |
Fulltext Views
1,165
PDF downloads
235




